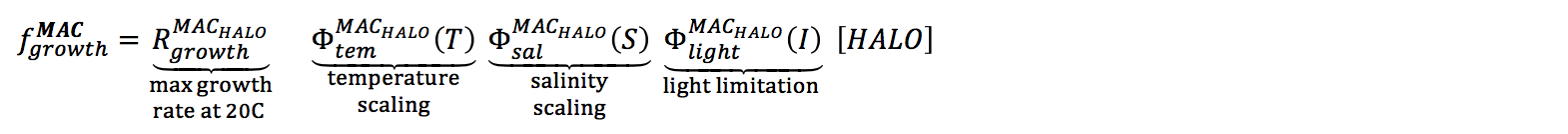Science Modules / Macrophyte
We approach the model by including a core state variable HALO, that is made up of above ground (AG) and below ground (BG); the former referring to the leaves, and the latter the cumulative mass of rhizomes and roots.
Goto Parameter Databaseaed2_macrophyte : Mass balance and functions related to the macrophyte model
The equation for seagrass biomass in a given cell is computed as shown in the below table, assuming photosynthesis, excretion, mortality and excretion. Nutrient uptake is not included in the simulation in SCERM v1 since Hillman et al. (1995) highlight that light, salinity and temperature were the dominant drivers, with some minor correlation with phosphate levels, however the stoichiometry is known and may be included in future iterations. The effective coverage area (i.e. Leaf Area Index, HALOLAI of the meadow) can be computed based on the scaling expression in Baird et al. (2016); although this is not directly used in the photosynthesis calculation, it is useful output for comparing with field data.
Growth is calculated in response to light, but also sensitive to salinity and temperature:
Photosynthesis-irradiance relationships for Halophila have been estimated by Ralph and Burchett (1995) who found photo-inhibition occurring at modest light intensities. The Steele (1962) equation is therefore suggested as the most appropriate. Light extinction can also occur over the meadow depth, although for Halophila this is assumed to be relatively small due to the small leaf structure. The salinity effect on photosynthesis has been reported by Ralph (1998b) and Hillman et al. (1995).
Respiration, excretion and mortality are also commonly simulated with typical first-order rate coefficients for each:
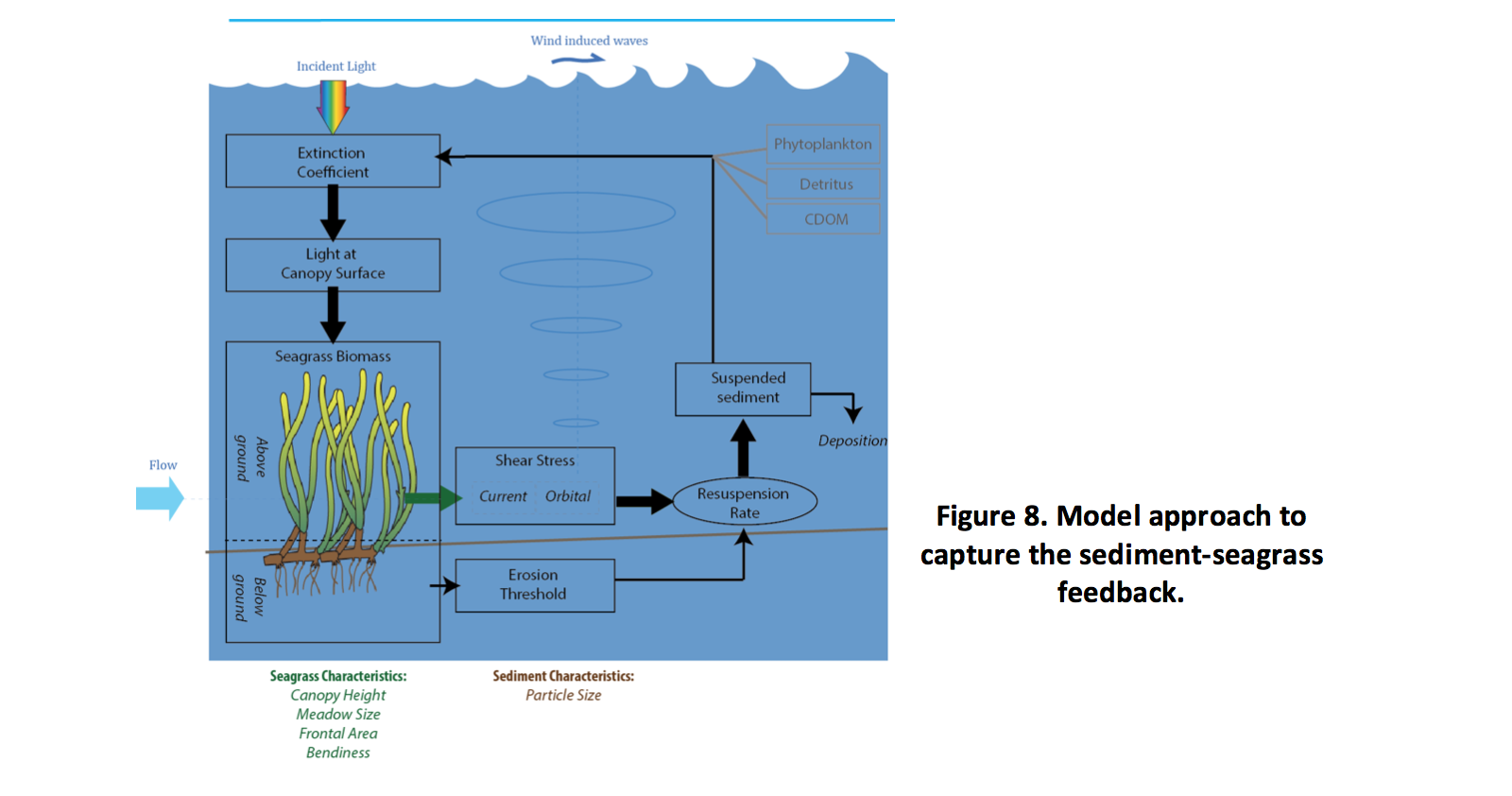
The seagrass-sediment-light (SSL) feedback has been identified as a potentially important driver determining meadow persistence (Adams et al., 2016), as depicted schematically for a numerical model in Figure 8. This requires the connection between sediment resuspension and seagrass presence to be made, however, the complete feedback loop has rarely been reported in aquatic models to date. The model implemented for SCERM focuses on Halophila and accounts for the feedback by including ability to simulate the link between: a) above ground biomass (l“DYÀE) and shear stress, b) below ground biomass (l“DYFE), and c) the amount of resuspension and light.
To simulate the SSL feedback, drag is increased in proportion with the above ground biomass:
where \(C_d\) is the base drag coefficient for a numerical cell based on its sediment material properties, and the above ground fraction is a user definable constant fraction of total biomass. The default value of \(C_{D_{bottom}}\)is stored in the hydrodynamic driver model and influences the local momentum budget; therefore the 2nd term of the RHS is passed to the host model calling AED2. The critical shear stress for resuspension is also increased based on the below ground biomass:
where \(\tau_0\) is critical shear stress for resuspension, computed based on a minimum value (reflecting bare sediment), and linear coefficient linked to biomass. As HI increases, the concentration of SS in the local domain will reduce, thereby improving the overall light climate. Note that in the model, presence or absence of HALO in a given cell can be configured be reading in an appropriate distribution map.
Variable Summary & Setup Options
| Variable Name | Description | Units | Variable Type | Core/Optional |
|---|---|---|---|---|
MAC_{Group} |
macrophyte group | $$mmol C\,m^{-2}$$ | - | core |
| Variable Name | Description | Units | Variable Type | Core/Optional |
|---|---|---|---|---|
MAC_PAR |
benthic light intensity | $$W\,m^{-2}$$ | - | - |
MAC_GPP |
benthic plant productivity | $$\,d^{-1}$$ | - | - |
MAC_P2R |
macrophyte p:r ratio | - | - | - |
MAC_MAC |
total macrophyte biomass | $$mmol C\,m^{-2}$$ | - | - |
MAC_LAI |
macrophyte leaf area density | $$m^{2}\,m^{-2}$$ | - | - |
MAC_MAC_ag |
total above ground macrophyte biomass | $$mmol C\,m^{-2}$$ | - | - |
MAC_bg |
total below ground macrophyte biomass | $$mmol C\,m^{-2}$$ | - | - |
| Parameter Name | Description | Units | Parameter Type | Default | Typical Range | Comment |
|---|---|---|---|---|---|---|
num_mphy |
number of macrophyte groups | - | integer | 0-5 | - | |
the_mphy |
list of ID's of groups in aed_phyto_pars.nml (len=num_phyto) | - | integer | 1 - 5 | - | |
n_zones |
total number of active material zones | - | integer | - | - | - |
active_zones |
Active material Zone ID's | - | array | - | - | - |
dbase |
link to mphy database | - | string | - | - | - |
simStaticBiomass |
switch to enable static biomass simulation | - | boolean | .false. | - | - |
simMacFeedback |
switch to enable macrophyte feedback simulation | - | boolean | .false. | - | - |
m_name |
name of macrphyte group | - | string | - | - | - |
m0 |
minimum above ground density of macrophyte | $$mmol C\,m^{-2}$$ | float | 2e-01 | - | - |
R_growth |
maximum growth rate at 20C | $$mmol C\,m^{-2}\,day^{-1}$$ | float | 2 | - | - |
fT_Method |
specific temperature limitation function of growth | - | integer | 0-1 | - | |
theta_growth |
Arrenhius temperature scaling for growth function | - | float | 1e+00 | - | - |
T_std |
standard temperature | deg C | float | 20 | - | - |
T_opt |
optimum temperature | deg C | float | 20 | - | - |
T_max |
maximum temperature | deg C | float | 20 | - | - |
lightModel |
type of light response function | - | integer | 0-1 | - | |
I_K |
half saturation constant for light limitation of growth | $$microE\,m^{-2}\,s^{-1}$$ | float | 180 | - | - |
I_S |
saturation light intensity used if lightModel=1 | $$microE\,m^{-2}\,s^{-1}$$ | float | 100 | - | - |
KeMAC |
specific attenuation coefficient | - | float | 4e-03 | - | - |
f_pr |
fraction of primary production lost to exudation | - | float | 3e-02 | - | - |
R_resp |
respiration loss rate at 20C | $$mmol C\,m^{-2}\,day^{-1}$$ | float | 2e-02 | - | - |
theta_resp |
Arrhenius temperature scaling factor for respiration | - | float | 1e+00 | - | - |
salTol |
type of salinity limitation function | - | float | - | - | |
S_bep |
salinity limitation value at miximum falinity s_maxsp | - | float | 1 | - | - |
S_maxsp |
maximum salinity | - | float | 36 | - | - |
S_opt |
optimal salinity | - | float | 1 | - | - |
K_CD |
constant relating macrophyte density to drag coefficient | $$mmol C\,m^{-3}\,day^{-1}$$ | float | - | - | |
f_bg |
fraction of primary production apportioned to below-ground biomass | $$mmol C\,m^{-3}\,day^{-1}$$ | float | - | - | |
k_omega |
Omega constant relating plant density to effective area of coverage | $$W\,m^{-2}$$ | float | - | - | |
Xcc |
internal C:chla ratio of the plant leaves | - | float | 50 | - | - |
K_N |
half saturation concentration of nitrogen | - | float | 2e+00 | - | - |
X_ncon |
constant internal nitrogen concentration | - | float | 2e-01 | - | - |
K_P |
half saturation concentration of phosphorus | - | float | 3e-01 | - | - |
X_pcon |
constant internal phosphorus concentration | $$kg\,m^{-3}$$ | float | 9e-03 | - | - |
An example nml block for the macrophyte module is shown below. NOTE: Users must supply a valid "aed2_macrophyte_pars.nml" file.
&aed2_macrophyte
num_mphy = 1
the_mphy = 3
n_zones = 4
active_zones = 2,3,4,5
simStaticBiomass = .true.
simMacFeedback = .false.
dbase = '../External/AED2/aed2_macrophyte_pars.nml'
/
Examples
Under construction
Publications & References
To be completed ...